Genomics and Applied Biology 2019, Vol.10, No.1, 1-9
2
In view of the fact that plant miRNAs is an important regulatory factor of plant growth and development,
secondary metabolism and stress resistance, the new generation of high-throughput sequencing technique was
used to analyze the miRNA of young bud, second leaf, open leaf and mature leaf of “Zijuan”, respectively.
miRNAs and its corresponding target genes in tea leaves of “Zijuan” were excavated, which provided theoretical
guidance for further study on the regulation of growth and development, secondary metabolism and stress
resistance of tea leaves by miRNAs.
1 Results and Analysis
1.1 High-throughput sequencing and assembly
In this study, 11,904,437 sequences were obtained from young buds of “Zijuan” by high-throughput sequencing,
10,532,877 sequences were obtained from the second leaves, 12,701,347 sequences were obtained from the open
leaves, and 12,042,833 sequences were obtained from the mature leaves. After removing the sequences with
inferior quality, contaminated joints, no 3’ connectors or inserts, and Reads containing ployA/T/G/C, 11,710,509,
10,354,353, 12,493,732, 11,832,854 Reads were obtained, respectively (Table 1).
Table 1 List of sequencing data quality
Samples
Raw data Clean date Proportion (%) Error rate (%)
Q20 (%)
Q30 (%)
Bud
11,904,437 11,710,509 98.37
0.01
98.77
97.86
Second-leaf
10,532,877 10,354,353 98.31
0.01
98.87
98.03
Open surface leaf 12,701,347 12,493,732 98.37
0.01
98.87
98.01
Mature-leaf
12,042,833 11,832,854 98.26
0.01
98.41
96.95
1.2 sRNA length screening and analysis
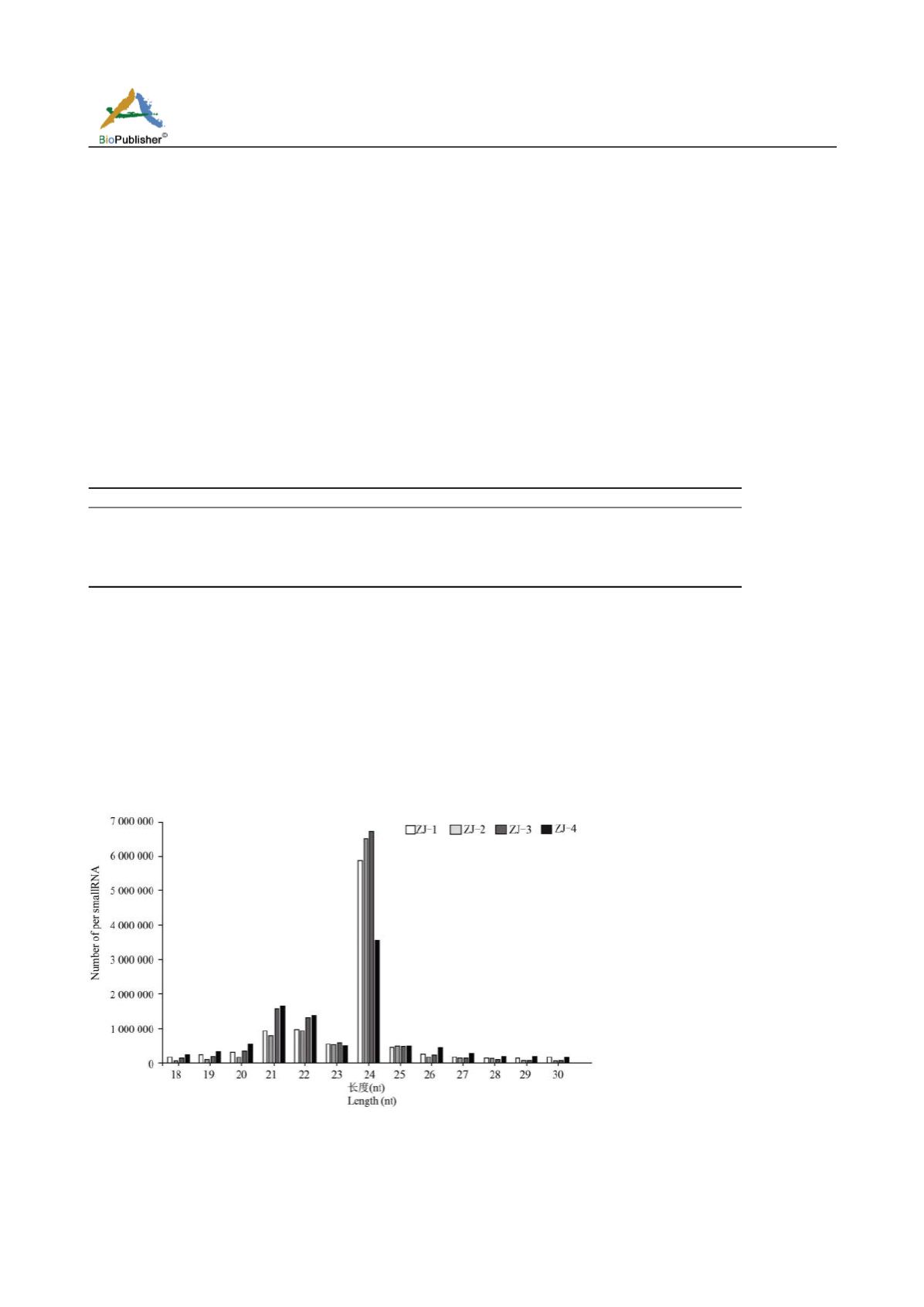
The Clean reads screening length ranged 18~30 nt, and sRNA was mainly distributed in the range of 21~24 nt, of
which 24 nt was the most (Figure 1). These sRNA distributions were consistent with other reports (Kwak et al.,
2009). Then, the screened sRNA was located on the corresponding reference sequence by bowtie software, and the
distribution of sRNA in the reference sequence was analyzed. The number of bud’s Clean reads mapped to
reference sequence was 5,410,363, the second leaf was 5,135,534, the open leaf was 5,944,894, and the mature
leaf was 3,831,890. Mapped sRNA was annotated with NCBI database (
) and Rfam
database. These annotated non-coding RNAs included known miRNA, new miRNAs, rRNAs, tRNAs, snRNAs,
snoRNAs, TAS gene (Table 2).
Figure 1 Abundance of small RNA sequence size
Note: ZJ-1: Bud; ZJ-2: Second-leaf; ZJ-3: Open surface leaf; ZJ-4: Mature-leaf
1.3 Identification of known miRNA and analysis of new miRNA gene
The Reads mapped onto the reference sequence were compared with the miRNA sequences of grape in miRBase,
and 126 known miRNAs were identified and divided into 26 families. Among these miRNA families, MIR169_2


