Basic HTML Version


Triticeae Genomics and Genetics 2012, Vol.3, No.3, 25
-
37
http://tgg.sophiapublisher.com
33
and rice largely share common syntenic pattern at
chromosomal level. We also suggest that the syntenic
conservation before and after polyploidization in
wheat, was still maintained for at least some wheat
chromosomes vis-à-vis the brachypodium and rice
chromosomes. Thus, the present study shed light on
the nature of chromosome evolution in polyploid
genomes such as wheat and other members of Triticeae.
3 Materials and Methods
3.1 Sequences used for synteny analysis
A total of 8 210 wheat ESTs (wESTs) belonging to all
the 21 chromosomes of wheat were downloaded from
GrainGenes-SQL database (http://wheat.pw.usda.gov/
cgi-bin/westsql/map_locus_rev.cgi). The bin-map
positions of these mapped ESTs were also retrieved
from the same database (http://wheat.pw.usda.gov/
NSF/progress_mapping.html; http://wheat.pw.usda.gov/
cgi-bin/westsql/map_locus.cgi).
A BLAST server (BrachyBlast) containing 8
×
seq-
uence of five brachypodium chromosomes/pseudo-
molecules (build; JGI v1.0 8
×
271 923 306 bp)
available on-line (http://www.brachypodium.org/; http://
www.brachybase.org/blast/) was used for BLAST
analysis. The alignment of each mapped wEST with
sequences of five different chromosomes/pseudomo-
lecules of brachypodium was carried out by using
independently each mapped wEST as a query
sequence in BLAST analysis.
3.2 Criteria used for sequence comparisons
In order to simplify the analysis and to ensure the
acceptable syntenic relationship between the chromo-
somes of wheat and brachypodium, we employed
integrative sequence alignment criteria for each
BLAST output with the following two parameters
(Salse et al., 2008; Kumar et al., 2009): (1) Cumulative
Identity Percentage (CIP) obtained using the following
formula: CIP = [
∑
Id of HSPs/AL]
×
100, where, Id
= identity; HSPs = length of high scoring segment
pairs and AL = alignment length between query and
subject sequences; and (2) Cumulative Alignment
Length Percentage (CALP): values of CALP were
calculated as follows: CALP = [AL/QL]
×
100, where,
QL is the length of query sequence.
While establishing synteny between wheat and bra-
chypodium genomes at chromosomal level, a value of
70% for CIP as well as CALP in each BLAST result
was considered stringent (Salse et al., 2008). In
several cases, BLAST output may often contain many
HSPs due to the occurrence of redundancy or
duplications in the genome. In such cases, we visually
scanned the BLAST output files to determine the
correct order of consecutive HSPs (in query sequence)
produced by alignment between query and subject
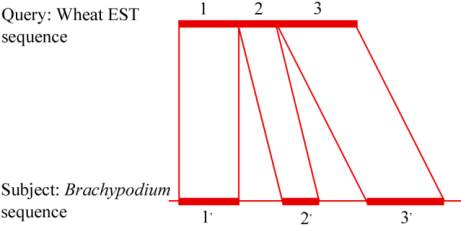
sequences (Figure 4).
Figure 4 BLASTN results, where segments 1, 2 and 3 are
consecutive segments in the query sequence (wEST) and their
match in the genomic sequence (1' 2' and 3') of brachypodium
are also consecutive, although in the brachypodium sequence,
there are two gaps, one between segments 1' and 2', and the
other between 2' and 3'; The segments (1, 2 and 3) of wheat
EST-sequence matching with segments (1' 2' and 3') of
brachypodium sequence make longest consecutive HSP in
correct order
3.3 Construction of consensus map of wheat genome
A map of all the seven homoeologous groups of
wheat was constructed on the basis of only those
wESTs that were bin-mapped to two or all the three
chromosomes of a homoeologous group and also
exhibited homology with sequences of brachypodium.
The methodology of constructing the consensus map
was described earlier in several studies (Gill et al.,
1996a, 1996b; Linkiewicz et al., 2004; Munkvold et
al., 2004). On the ordered deletion bins, ESTs that
detected duplicate (paralogous) loci on two or three
homoeologous chromosomes were grouped according
to their bin positions on each chromosome and
consensus bins were defined by ESTs sharing a
common mapping pattern in the same group. For
example, wEST BE404868 (showing homology with
brachypodium chromosome 2) mapping to the deletion
bins 3AS4
-
0.45
-
1.00, 3BS1
-
0.33
-
0.57 and 3DS3
-
Triticeae Genomics and Genetics Provisional Publishing

