Rice Genomics and Genetics 2015, Vol.6, No.1, 1-8
6
sequence data. The phylogenetic study of the
matK
nucleotide sequences of ten aromatic rice genotypes
including related wild species and other grasses
family under Poaceae concludes that the genus
Oryza
was divided into two main clades and evolved
from completely different group of grass families.
Aromatic rice might be evolved from
Oryza sativa
(japonica and Indica group),
Oryza glaberrima
or
Oryza rufipogal
because of its low bootstrap value
support.
Oryza brachyantha
has high affinity for
grasses and should be treated as a progenitor for
wild
Oryza
species. The study will also address the
issues of optimal number of nucleotides essential for
robust phylogenies and the consequences of utilizing
different segments of the gene.
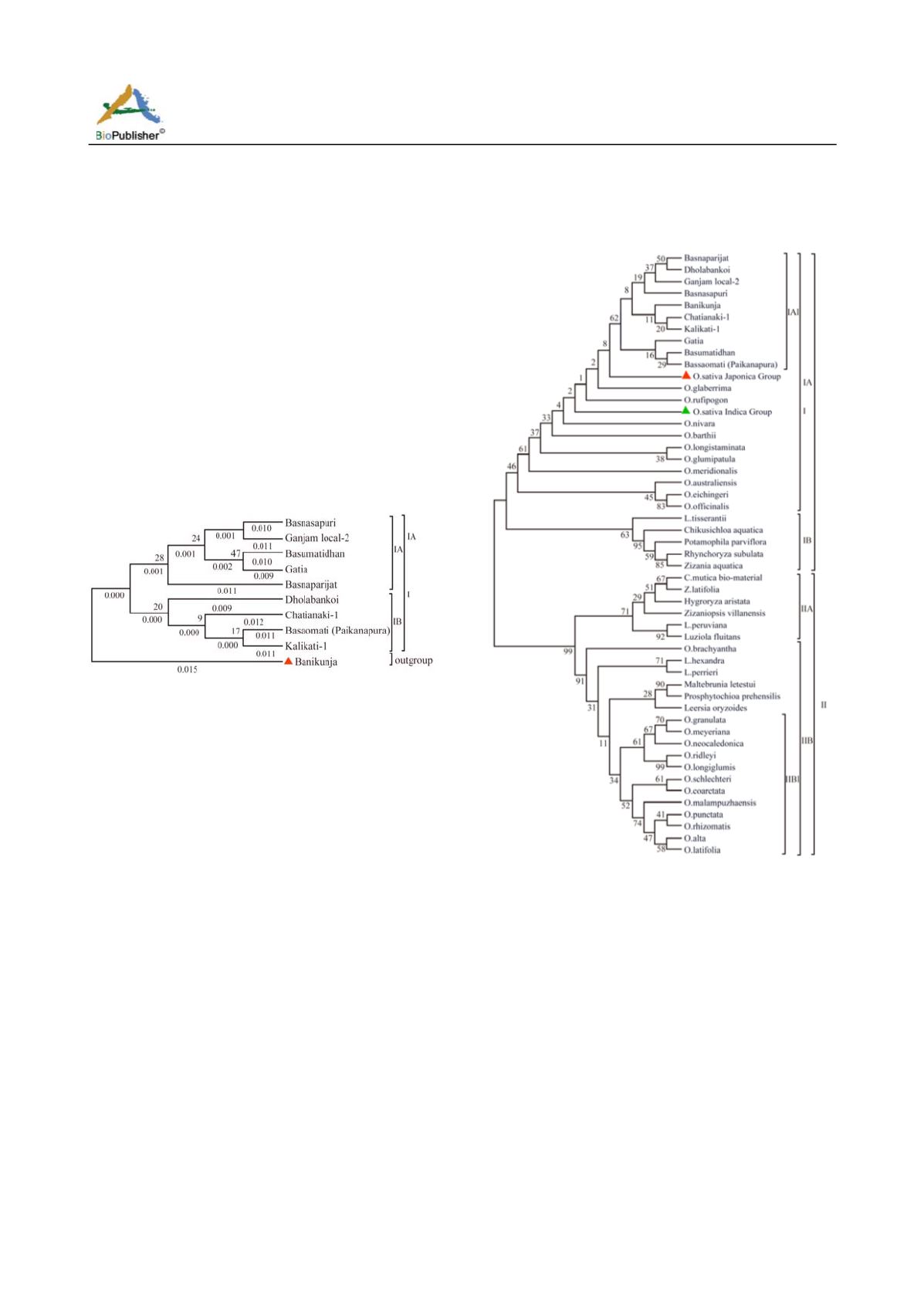
Figure 3 Phylogenetic tree based on nucleotide sequences of
10 aromatic rice cultivars constructed by neighbor joining
method
Note: Out group indicates in red mark. Black bold fond on
upper side of the tree node indicates bootstrap values in per
cent. Values in black fond below the tree root indicates the
divergence time
Materials and Methods
Genotypes
Ten promising upland as well as lowland cultivars of
indigenous aromatic rice (
Oryza sativa
L.) viz.
‘Basnasapuri’, ‘Basnaparijat, ‘Basumati dhan’,
‘Banikunja’, ‘Basaomati (Paikanapura)’, ‘Chatianaki - 1’,
‘Dhoiabankoi’, ‘Ganjam local - 2’, ‘Kalikati - 1’, and
‘Gatia’ were collected from the Rice Research Station,
Orissa university of Agriculture and Technology,
Bhubaneswar, Odisha, India and used in the present
study.
DNA extraction and PCR amplification
Total cellular DNAs were extracted from young
leaftissues of ten aromatic rice genotypes using
modified CTAB method (Edwards et al., 1991) and
purified the DNA of each genotype was subjected
for PCR amplification using overlapping oligos viz.
matK
F1: 5’TAATTAAGAGGATTCACC AG 3’ and
matK
R1: 5‘ATGCAACACCCTGTTCTGAC3’ (Merck
Bioscience, India) by examining the previous study
Figure 4 Phylogenetic tree based on the alignment of
nucleotide sequences ofaromatic rice cultivars and its wild
relatives along with progenitors grasses constructed by
neighbor joining method.
Note: Common rice indicates in red and green mark. Black bold
on rice (Ge et al., 1999).PCR amplification was
carried out with the template DNA (25~50 ng), 2.5
µl 10X PCR assay buffer (Merck Bioscience,
India),1.5 µl each of 10 mM dNTPs (M/S Merck
Bioscience, India), 1 µl of 5 µM forward primer, 1
µl of 5 µM primer and 1 µl of 1U
Taq
DNA
polymerase (Merck Bioscience). M/s Peqlab, 96
universal gradient thermal cycler was used for


