Basic HTML Version


Molecular Plant Breeding 2013, Vol.4, No.38, 297
-
303
http://mpb.sophiapublisher.com
300
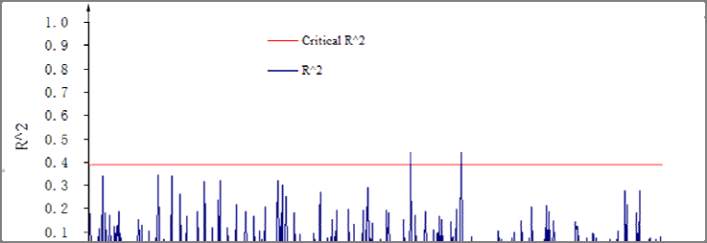
Figure 3 Linkage disequilibrium (
r
2
) as a function of physical distance (Mb) among 141 markers
Note: The red line represents critical values of
r
2
Mather et al (2007) also reported that the extent of
linkage disequilibrium differs in significant way
between domesticated Asian rice and
O. rufipogon
,
both in genome wide LD and in targeted genomic
regions, higher LD was found in
O. sativa
variety
groups. In the presence of high LD, lower marker
density is required for a target region with greater
potential for detecting markers strongly associated
with the target gene polymorphism, even if distant
physically (Agrama et al., 2007), whole-genome-scan
association study is feasible in this case.
The success of association mapping efforts depends
on the possibilities of separating LD due to linkage
from LD due to other causes. Factors such as the
mating system, the recombination rate, population
structure, population history, genetic drift, directional
selection, and gene fixation at different rates on
different chromosome regions can all affect the
patterns of LD (Gaut and Long, 2003). In general, LD
decay with distance occurs at a much slower rate in
self pollinated plants, such as
Arabidopsis
, rice, barley,
durum wheat, and sorghum, than in outcrossing
species. In this study, LD decayed faster than 3 MB
bin, Rakshit et al. (2007) reported an LD decay of
~50Kb in
indica
and of ~5 Kb in
O.rufipogon
, which
was significantly smaller as compared with 3 MB in
this study, a fast decay in the genome, however, is an
inference of high resolution for association analysis.
However, two pairs of markers RM288 and RM464
(11.98758 Mb), RM215 and RM464 (14.61396 Mb)
all were located on chromosome 9 but at relatively far
distance showed significant linkage. The same
phenomenon also reported by Skot (2005)
in the
natural population of perennial ryegrass at a
genome-wide scale using AFLP genetic markers in
which the majority of the linked pairs were in
significant LD within genetic distance of 4.37 cM
with
r
2
=0.12, but two pairs were more than 20cM
apart. These Loci might co-segregated through long
term selection, as natural or artificial selection of
varied trait favors co-inheritance of those loci, which
was another factor cause linkage equilibrium.
Selection for or against a phenotype controlled by
alleles at two unlinked loci that show epistatic
interaction may result in LD despite the fact that the
loci are not physically linked (Palaisk et al., 2003).
3 Conclusion
Significant LD was detected across the genome of the
184 rice genotypes, selfing in
O. sativa
species and
existing of population structure in the rice panel might
be the major factors of creating high LD. However, as
a high LD indicated a lower marker density would be
enough to detect markers strongly associated with the
trait of interest, and whole-genome-scan association
study with currently used 141 SSR markers could be
feasible in this case. The majority of the linked pairs
were in significant LD with
r
2
=0.3886, LD decayed
faster than 3 MB bin in this mapping population, a
comparatively higher resolution for association
analysis with the 141 SSR markers would be
expected.

