Basic HTML Version




Molecular Pathogens
MP2011, Vol.2, No.2
http://mp.sophiapublisher.com
- 13 -
coat protein sequence) with an amplification product
of 374 bp was obtained (Figure 5). The sequence was
deposited in GenBank under accession number (Acc#
GQ288368). This band was successfully amplified
from RNA extracted from both symptomatic tissues
and purified viruses (Figure 5). The sequence of this
fragment showed one ORF within the FMV coat
protein (
CP
) gene. Sequence alignment showed that
the site of this fragment was about 1-146 codons after
the starting codon, AUG of the FMV coat protein gene.
An ORF that could encode a polypeptide of 124
amino acids was detected. This deduced polypeptide
contains 15 strongly basic, 14 strongly acidic, 35
hydrophobic, and 33 polar amino acids. The
calculated molecular mass of the putative polypeptide
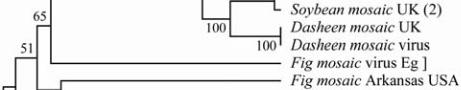
is 13.67 kDa. Phylogenetic tree construction was
based on the deduced amino acid sequences for the
obtained CP with other coat protein genes for 14 Fig
mosaic viruses, as depicted in Figure 6 and Figure 7.
The neighbor-joining distance analysis with maximum
sequence difference of 1.2 and the topology yielded
four distinct lineages that were similar to the Arkansas
mosaic virus (Acc# FJ769161, with identity 65%) and
to the Italian mosaic virus (Acc# FM864225, with
identity 65%).
Figure 7 Phylogenetic analysis based on the deduced amino
acid sequences showing the genetic
relationship between the
NIb
genes of FMV with those of
selected other viruses
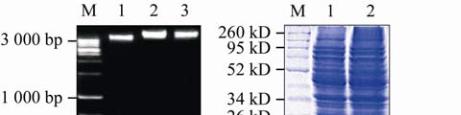
1.5 Recombinant coat protein and SDS analysis
The recombinant cells were harvested and hydrolyzed;
and the recombinant protein was separated on 12%
SDS-PAGE. The protein with molecular weight of
13.7 kDa was presented only in the recombinant
bacterial cells compared with the non recombinant
ones (Figure 8).
Figure 8 Agarose gel electrophoresis and SDS-PAGE of the
cloning coat protein gene and the
Fig Mosaic Virus
coat protein
gene
Note: A: Agarose gel electrophoresis (1.5%) showing
confirmation of the cloning of the coat protein gene; M: 1 kb
Ladder DNA Marker; 1: Uncharged plasmid; 2,3: Charged
plasmid (recombinant plasmid); B: 12% SDS-PAGE for the
E.coli
empty and the recombinant
E.coli
contains the
Fig
Mosaic Virus
coat protein gene; M: Protein Marker; 1:
Non-induced sample; 2: Induced sample contains the
recombinant coat protein at 13.7 kD
2 Discussion
According to FAO (2008), the Mediterranean basin
area is known to produce 80% of global production of
fig. Egypt provides 27%, Turkey 11% and Europe
15%, in addition to the other countries. Egypt, Turkey,
Iran, Algeria and Morocco were considered the top
five fig-producing countries in the world. Fig mosaic
disease (FMD) is an economically important disease
that occurs naturally, wherever the common edible fig
(
Ficus carica
L.) grows. It was observed in 14
different geographical origins of the world: Spain,
England, Albania, Cyprus, Greece, Turkey, Israel,
Yemen, Egypt, Tunisia, Algeria, Morocco, Italy and
California (Martelli et al., 1993 and Ahn et al., 1996).
Among important viruses that caused devastating
losses by reducing either the yield and/or quality of fig
fruits is
fig mosaic virus
(FMV). For this purpose, the
present investigation aimed to identify unidentified
isolate of this virus based on different biological,
serological and molecular tools, in order to provide a
powerful diagnostic tool for early detection of FMV in
infected tissues. In the present study, symptomatology
identification of the FMV causes symptoms on both
leaves and fruits. The mosaic spots were distinctly
yellow on leaves, contrasting with normal green color

