Basic HTML Version



Genomics and Applied Biology, 2011, Vol.2 No.5
http://gab.sophiapublisher.com
- 30 -
thus, developing of lodging-resistant cultivar has been
a longstanding challenge. The author developed a
dwarf Koshihikari-type cultivar, ‘Hikarishinseiki’ (Tomita,
2009; rice cultivar No.12273, Ministry of Agriculture,
Forestry and Fisheries of Japan) using a dwarf cultivar
line with the heading time same as Koshihikari-type,
which was selected by intercrossing the F
4
of Kanto
No.79 (early-heading mutant line derived from Koshi-
hikari) and Jukkoku (a cultivar with a semi-dwarf gene,
sd1
), as the maternal parent, and backcrossed it with
Koshihikari 8 times. Having over 99.8% background of
the Koshihikari genome, except for
sd1
, Hikarishinseiki
would be the first cultivar to be registered as a Koshi-
hikari-type dwarf with
sd1
in Japan (Tomita, 2009).
The dwarf feature of Hikarishinseiki is derived from
the semidwarf gene
sd1
.
sd1
is a defective gene of
GA20ox
-
2
, which encodes a defective C20
-
oxidase in
the gibberellin (GA) biosynthesis pathway (GA 20
-
oxidase,
OsGA20ox2
) (Monna et al., 2002; Spielmeyer et
al., 2002; Sasaki et al., 2002; Ashikari et al., 2002).
However, it has yet to be known whether the
sd1
gene
possessed by Hikarishinseiki is transcribed. Subsequently,
expression analysis was conducted by employing the
RT-PCR method and
Pma
CI restriction enzyme digestion
in order to clarify whether or not the
sd1
gene carried
by Hikarishinseiki is transcribed.
An increasing number of private agricultural producers
across Japan have spontaneously started to grow Hika-
rishinseiki as a typhoon-resistant, easy-to-grow cultivar
instead of Koshihikari. As the recommended cultivars
become widespread, there will become a growing need
for information that rewards producers for their efforts.
Here, we analyzed the results of 24 performance tests
aimed at determining recommended cultivars, carried
out over a period of two years from 2006 to 2007.
1 Results
1.1 Transcription of
sd1
, defective allele of
GA20ox-2
We performed expression analysis of the
sd1
gene,
which is introduced in the dwarf Koshihikari-type
paddy rice variety ‘Hikarishinseiki’. The two alleles at
the
sd1/OsGA20ox2
locus on chromosome 1 of each
line, Koshihikari and Hikarishinseiki, were distinguished
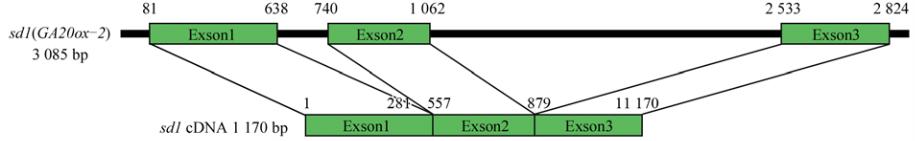
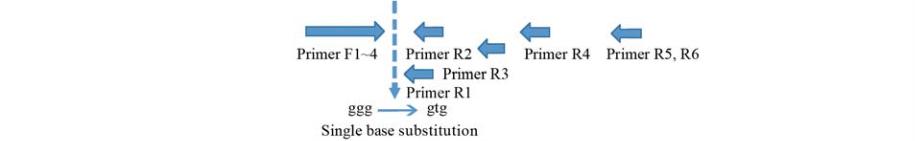
by RT-PCR amplification of the first exon followed by
digestion with
Pma
CI (Figure 1; Figure 2). A 779 bp
fragment, employing one type of upper primer (F1)
designed from the 1
st
exon and one type of down-
stream primer (R4) designed from the range of the 2
nd
to 3
rd
exon, contained only the exon. Moreover, 1,316 bp,
1,304 bp, 1,400 bp, and 1,366 bp fragments employing
the four upper primers (F1~F4) designed from the 1
st
exon and the downstream primer (R6) designed from
the 3
rd
exon, contained the 2
nd
intron. Lastly, 1,428 bp,
1,416 bp, and 1,478 bp fragments, obtained using three
upstream primers (F1~F3) designed from the 1
st
exon
and the RT primer (R5), contained both the 1
st
intron
and 2
nd
intron. The target fragment was detected in
these eight combinations of primers.
Figure 1 PCR primers designed for amplification of cDNA derived from
sd1
(
GA20ox
-
2
) transcript

